Sensitivity and specificity
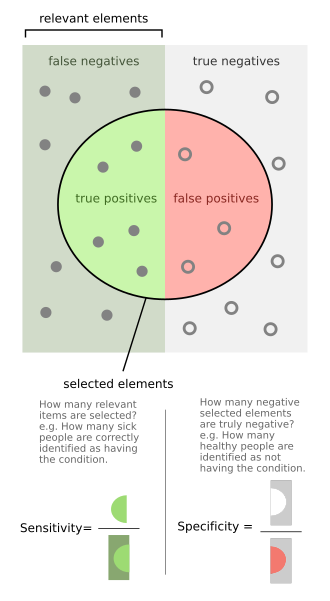
In medicine and statistics, sensitivity and specificity mathematically describe the accuracy of a test that reports the presence or absence of a medical condition. If individuals who have the condition are considered "positive" and those who do not are considered "negative", then sensitivity is a measure of how well a test can identify true positives and specificity is a measure of how well a test can identify true negatives:
- Sensitivity (true positive rate) is the probability of a positive test result, conditioned on the individual truly being positive.
- Specificity (true negative rate) is the probability of a negative test result, conditioned on the individual truly being negative.
If the true status of the condition cannot be known, sensitivity and specificity can be defined relative to a "gold standard test" which is assumed correct. For all testing, both diagnoses and screening, there is usually a trade-off between sensitivity and specificity, such that higher sensitivities will mean lower specificities and vice versa.
A test which reliably detects the presence of a condition, resulting in a high number of true positives and low number of false negatives, will have a high sensitivity. This is especially important when the consequence of failing to treat the condition is serious and/or the treatment is very effective and has minimal side effects.
A test which reliably excludes individuals who do not have the condition, resulting in a high number of true negatives and low number of false positives, will have a high specificity. This is especially important when people who are identified as having a condition may be subjected to more testing, expense, stigma, anxiety, etc.

The terms "sensitivity" and "specificity" were introduced by American biostatistician Jacob Yerushalmy in 1947.[1]
There are different definitions within laboratory quality control, wherein "analytical sensitivity" is defined as the smallest amount of substance in a sample that can accurately be measured by an assay (synonymously to detection limit), and "analytical specificity" is defined as the ability of an assay to measure one particular organism or substance, rather than others.[2] However, this article deals with diagnostic sensitivity and specificity as defined at top.
Application to screening study
[edit]Imagine a study evaluating a test that screens people for a disease. Each person taking the test either has or does not have the disease. The test outcome can be positive (classifying the person as having the disease) or negative (classifying the person as not having the disease). The test results for each subject may or may not match the subject's actual status. In that setting:
- True positive: Sick people correctly identified as sick
- False positive: Healthy people incorrectly identified as sick
- True negative: Healthy people correctly identified as healthy
- False negative: Sick people incorrectly identified as healthy
After getting the numbers of true positives, false positives, true negatives, and false negatives, the sensitivity and specificity for the test can be calculated. If it turns out that the sensitivity is high then any person who has the disease is likely to be classified as positive by the test. On the other hand, if the specificity is high, any person who does not have the disease is likely to be classified as negative by the test. An NIH web site has a discussion of how these ratios are calculated.[3]
Definition
[edit]Sensitivity
[edit]Consider the example of a medical test for diagnosing a condition. Sensitivity (sometimes also named the detection rate in a clinical setting) refers to the test's ability to correctly detect ill patients out of those who do have the condition.[4] Mathematically, this can be expressed as:
A negative result in a test with high sensitivity can be useful for "ruling out" disease,[4] since it rarely misdiagnoses those who do have the disease. A test with 100% sensitivity will recognize all patients with the disease by testing positive. In this case, a negative test result would definitively rule out the presence of the disease in a patient. However, a positive result in a test with high sensitivity is not necessarily useful for "ruling in" disease. Suppose a 'bogus' test kit is designed to always give a positive reading. When used on diseased patients, all patients test positive, giving the test 100% sensitivity. However, sensitivity does not take into account false positives. The bogus test also returns positive on all healthy patients, giving it a false positive rate of 100%, rendering it useless for detecting or "ruling in" the disease.
The calculation of sensitivity does not take into account indeterminate test results. If a test cannot be repeated, indeterminate samples either should be excluded from the analysis (the number of exclusions should be stated when quoting sensitivity) or can be treated as false negatives (which gives the worst-case value for sensitivity and may therefore underestimate it).
A test with a higher sensitivity has a lower type II error rate.
Specificity
[edit]Consider the example of a medical test for diagnosing a disease. Specificity refers to the test's ability to correctly reject healthy patients without a condition. Mathematically, this can be written as:
A positive result in a test with high specificity can be useful for "ruling in" disease, since the test rarely gives positive results in healthy patients.[5] A test with 100% specificity will recognize all patients without the disease by testing negative, so a positive test result would definitively rule in the presence of the disease. However, a negative result from a test with high specificity is not necessarily useful for "ruling out" disease. For example, a test that always returns a negative test result will have a specificity of 100% because specificity does not consider false negatives. A test like that would return negative for patients with the disease, making it useless for "ruling out" the disease.
A test with a higher specificity has a lower type I error rate.
Graphical illustration
[edit]-
A graphical illustration of sensitivity and specificity
The above graphical illustration is meant to show the relationship between sensitivity and specificity. The black, dotted line in the center of the graph is where the sensitivity and specificity are the same. As one moves to the left of the black dotted line, the sensitivity increases, reaching its maximum value of 100% at line A, and the specificity decreases. The sensitivity at line A is 100% because at that point there are zero false negatives, meaning that all the negative test results are true negatives. When moving to the right, the opposite applies, the specificity increases until it reaches the B line and becomes 100% and the sensitivity decreases. The specificity at line B is 100% because the number of false positives is zero at that line, meaning all the positive test results are true positives.
-
High sensitivity and low specificity
-
Low sensitivity and high specificity
The middle solid line in both figures above that show the level of sensitivity and specificity is the test cutoff point. As previously described, moving this line results in a trade-off between the level of sensitivity and specificity. The left-hand side of this line contains the data points that tests below the cut off point and are considered negative (the blue dots indicate the False Negatives (FN), the white dots True Negatives (TN)). The right-hand side of the line shows the data points that tests above the cut off point and are considered positive (red dots indicate False Positives (FP)). Each side contains 40 data points.
For the figure that shows high sensitivity and low specificity, there are 3 FN and 8 FP. Using the fact that positive results = true positives (TP) + FP, we get TP = positive results - FP, or TP = 40 - 8 = 32. The number of sick people in the data set is equal to TP + FN, or 32 + 3 = 35. The sensitivity is therefore 32 / 35 = 91.4%. Using the same method, we get TN = 40 - 3 = 37, and the number of healthy people 37 + 8 = 45, which results in a specificity of 37 / 45 = 82.2 %.
For the figure that shows low sensitivity and high specificity, there are 8 FN and 3 FP. Using the same method as the previous figure, we get TP = 40 - 3 = 37. The number of sick people is 37 + 8 = 45, which gives a sensitivity of 37 / 45 = 82.2 %. There are 40 - 8 = 32 TN. The specificity therefore comes out to 32 / 35 = 91.4%.
-
A test result with 100 percent sensitivity
-
A test result with 100 percent specificity
The red dot indicates the patient with the medical condition. The red background indicates the area where the test predicts the data point to be positive. The true positive in this figure is 6, and false negatives of 0 (because all positive condition is correctly predicted as positive). Therefore, the sensitivity is 100% (from 6 / (6 + 0)). This situation is also illustrated in the previous figure where the dotted line is at position A (the left-hand side is predicted as negative by the model, the right-hand side is predicted as positive by the model). When the dotted line, test cut-off line, is at position A, the test correctly predicts all the population of the true positive class, but it will fail to correctly identify the data point from the true negative class.
Similar to the previously explained figure, the red dot indicates the patient with the medical condition. However, in this case, the green background indicates that the test predicts that all patients are free of the medical condition. The number of data point that is true negative is then 26, and the number of false positives is 0. This result in 100% specificity (from 26 / (26 + 0)). Therefore, sensitivity or specificity alone cannot be used to measure the performance of the test.
Medical usage
[edit]In medical diagnosis, test sensitivity is the ability of a test to correctly identify those with the disease (true positive rate), whereas test specificity is the ability of the test to correctly identify those without the disease (true negative rate). If 100 patients known to have a disease were tested, and 43 test positive, then the test has 43% sensitivity. If 100 with no disease are tested and 96 return a completely negative result, then the test has 96% specificity. Sensitivity and specificity are prevalence-independent test characteristics, as their values are intrinsic to the test and do not depend on the disease prevalence in the population of interest.[6] Positive and negative predictive values, but not sensitivity or specificity, are values influenced by the prevalence of disease in the population that is being tested. These concepts are illustrated graphically in this applet Bayesian clinical diagnostic model which show the positive and negative predictive values as a function of the prevalence, sensitivity and specificity.
Misconceptions
[edit]It is often claimed that a highly specific test is effective at ruling in a disease when positive, while a highly sensitive test is deemed effective at ruling out a disease when negative.[7][8] This has led to the widely used mnemonics SPPIN and SNNOUT, according to which a highly specific test, when positive, rules in disease (SP-P-IN), and a highly sensitive test, when negative, rules out disease (SN-N-OUT). Both rules of thumb are, however, inferentially misleading, as the diagnostic power of any test is determined by the prevalence of the condition being tested, the test's sensitivity and its specificity.[9][10][11] The SNNOUT mnemonic has some validity when the prevalence of the condition in question is extremely low in the tested sample.
The tradeoff between specificity and sensitivity is explored in ROC analysis as a trade off between TPR and FPR (that is, recall and fallout).[12] Giving them equal weight optimizes informedness = specificity + sensitivity − 1 = TPR − FPR, the magnitude of which gives the probability of an informed decision between the two classes (> 0 represents appropriate use of information, 0 represents chance-level performance, < 0 represents perverse use of information).[13]
Sensitivity index
[edit]The sensitivity index or d′ (pronounced "dee-prime") is a statistic used in signal detection theory. It provides the separation between the means of the signal and the noise distributions, compared against the standard deviation of the noise distribution. For normally distributed signal and noise with mean and standard deviations and , and and , respectively, d′ is defined as:
An estimate of d′ can be also found from measurements of the hit rate and false-alarm rate. It is calculated as:
- d′ = Z(hit rate) − Z(false alarm rate),[15]
where function Z(p), p ∈ [0, 1], is the inverse of the cumulative Gaussian distribution.
d′ is a dimensionless statistic. A higher d′ indicates that the signal can be more readily detected.
Confusion matrix
[edit]The relationship between sensitivity, specificity, and similar terms can be understood using the following table. Consider a group with P positive instances and N negative instances of some condition. The four outcomes can be formulated in a 2×2 contingency table or confusion matrix, as well as derivations of several metrics using the four outcomes, as follows:
| Predicted condition | Sources: [16][17][18][19][20][21][22][23] | ||||
| Total population = P + N |
Predicted positive (PP) | Predicted negative (PN) | Informedness, bookmaker informedness (BM) = TPR + TNR − 1 |
Prevalence threshold (PT) = √TPR × FPR - FPR/TPR - FPR | |
Actual condition
|
Positive (P) [a] | True positive (TP), hit[b] |
False negative (FN), miss, underestimation |
True positive rate (TPR), recall, sensitivity (SEN), probability of detection, hit rate, power = TP/P = 1 − FNR |
False negative rate (FNR), miss rate type II error [c] = FN/P = 1 − TPR |
| Negative (N)[d] | False positive (FP), false alarm, overestimation |
True negative (TN), correct rejection[e] |
False positive rate (FPR), probability of false alarm, fall-out type I error [f] = FP/N = 1 − TNR |
True negative rate (TNR), specificity (SPC), selectivity = TN/N = 1 − FPR | |
| Prevalence = P/P + N |
Positive predictive value (PPV), precision = TP/PP = 1 − FDR |
False omission rate (FOR) = FN/PN = 1 − NPV |
Positive likelihood ratio (LR+) = TPR/FPR |
Negative likelihood ratio (LR−) = FNR/TNR | |
| Accuracy (ACC) = TP + TN/P + N |
False discovery rate (FDR) = FP/PP = 1 − PPV |
Negative predictive value (NPV) = TN/PN = 1 − FOR |
Markedness (MK), deltaP (Δp) = PPV + NPV − 1 |
Diagnostic odds ratio (DOR) = LR+/LR− | |
| Balanced accuracy (BA) = TPR + TNR/2 |
F1 score = 2 PPV × TPR/PPV + TPR = 2 TP/2 TP + FP + FN |
Fowlkes–Mallows index (FM) = √PPV × TPR |
Matthews correlation coefficient (MCC) = √TPR × TNR × PPV × NPV - √FNR × FPR × FOR × FDR |
Threat score (TS), critical success index (CSI), Jaccard index = TP/TP + FN + FP | |
- ^ the number of real positive cases in the data
- ^ A test result that correctly indicates the presence of a condition or characteristic
- ^ Type II error: A test result which wrongly indicates that a particular condition or attribute is absent
- ^ the number of real negative cases in the data
- ^ A test result that correctly indicates the absence of a condition or characteristic
- ^ Type I error: A test result which wrongly indicates that a particular condition or attribute is present
- A worked example
- A diagnostic test with sensitivity 67% and specificity 91% is applied to 2030 people to look for a disorder with a population prevalence of 1.48%
| Fecal occult blood screen test outcome | |||||
| Total population (pop.) = 2030 |
Test outcome positive | Test outcome negative | Accuracy (ACC) = (TP + TN) / pop.
= (20 + 1820) / 2030 ≈ 90.64% |
F1 score = 2 × precision × recall/precision + recall
≈ 0.174 | |
| Patients with bowel cancer (as confirmed on endoscopy) |
Actual condition positive (AP) = 30 (2030 × 1.48%) |
True positive (TP) = 20 (2030 × 1.48% × 67%) |
False negative (FN) = 10 (2030 × 1.48% × (100% − 67%)) |
True positive rate (TPR), recall, sensitivity = TP / AP
= 20 / 30 ≈ 66.7% |
False negative rate (FNR), miss rate = FN / AP
= 10 / 30 ≈ 33.3% |
| Actual condition negative (AN) = 2000 (2030 × (100% − 1.48%)) |
False positive (FP) = 180 (2030 × (100% − 1.48%) × (100% − 91%)) |
True negative (TN) = 1820 (2030 × (100% − 1.48%) × 91%) |
False positive rate (FPR), fall-out, probability of false alarm = FP / AN
= 180 / 2000 = 9.0% |
Specificity, selectivity, true negative rate (TNR) = TN / AN
= 1820 / 2000 = 91% | |
| Prevalence = AP / pop.
= 30 / 2030 ≈ 1.48% |
Positive predictive value (PPV), precision = TP / (TP + FP)
= 20 / (20 + 180) = 10% |
False omission rate (FOR) = FN / (FN + TN)
= 10 / (10 + 1820) ≈ 0.55% |
Positive likelihood ratio (LR+) = TPR/FPR
= (20 / 30) / (180 / 2000) ≈ 7.41 |
Negative likelihood ratio (LR−) = FNR/TNR
= (10 / 30) / (1820 / 2000) ≈ 0.366 | |
| False discovery rate (FDR) = FP / (TP + FP)
= 180 / (20 + 180) = 90.0% |
Negative predictive value (NPV) = TN / (FN + TN)
= 1820 / (10 + 1820) ≈ 99.45% |
Diagnostic odds ratio (DOR) = LR+/LR−
≈ 20.2 | |||
Related calculations
- False positive rate (α) = type I error = 1 − specificity = FP / (FP + TN) = 180 / (180 + 1820) = 9%
- False negative rate (β) = type II error = 1 − sensitivity = FN / (TP + FN) = 10 / (20 + 10) ≈ 33%
- Power = sensitivity = 1 − β
- Positive likelihood ratio = sensitivity / (1 − specificity) ≈ 0.67 / (1 − 0.91) ≈ 7.4
- Negative likelihood ratio = (1 − sensitivity) / specificity ≈ (1 − 0.67) / 0.91 ≈ 0.37
- Prevalence threshold = ≈ 0.2686 ≈ 26.9%
This hypothetical screening test (fecal occult blood test) correctly identified two-thirds (66.7%) of patients with colorectal cancer.[a] Unfortunately, factoring in prevalence rates reveals that this hypothetical test has a high false positive rate, and it does not reliably identify colorectal cancer in the overall population of asymptomatic people (PPV = 10%).
On the other hand, this hypothetical test demonstrates very accurate detection of cancer-free individuals (NPV ≈ 99.5%). Therefore, when used for routine colorectal cancer screening with asymptomatic adults, a negative result supplies important data for the patient and doctor, such as ruling out cancer as the cause of gastrointestinal symptoms or reassuring patients worried about developing colorectal cancer.
Estimation of errors in quoted sensitivity or specificity
[edit]Sensitivity and specificity values alone may be highly misleading. The 'worst-case' sensitivity or specificity must be calculated in order to avoid reliance on experiments with few results. For example, a particular test may easily show 100% sensitivity if tested against the gold standard four times, but a single additional test against the gold standard that gave a poor result would imply a sensitivity of only 80%. A common way to do this is to state the binomial proportion confidence interval, often calculated using a Wilson score interval.
Confidence intervals for sensitivity and specificity can be calculated, giving the range of values within which the correct value lies at a given confidence level (e.g., 95%).[26]
Terminology in information retrieval
[edit]In information retrieval, the positive predictive value is called precision, and sensitivity is called recall. Unlike the Specificity vs Sensitivity tradeoff, these measures are both independent of the number of true negatives, which is generally unknown and much larger than the actual numbers of relevant and retrieved documents. This assumption of very large numbers of true negatives versus positives is rare in other applications.[13]
The F-score can be used as a single measure of performance of the test for the positive class. The F-score is the harmonic mean of precision and recall:
In the traditional language of statistical hypothesis testing, the sensitivity of a test is called the statistical power of the test, although the word power in that context has a more general usage that is not applicable in the present context. A sensitive test will have fewer Type II errors.
Terminology in genome analysis
[edit]Similarly to the domain of information retrieval, in the research area of gene prediction, the number of true negatives (non-genes) in genomic sequences is generally unknown and much larger than the actual number of genes (true positives). The convenient and intuitively understood term specificity in this research area has been frequently used with the mathematical formula for precision and recall as defined in biostatistics. The pair of thus defined specificity (as positive predictive value) and sensitivity (true positive rate) represent major parameters characterizing the accuracy of gene prediction algorithms. [27] [28] [29] [30] Conversely, the term specificity in a sense of true negative rate would have little, if any, application in the genome analysis research area.
See also
[edit]Notes
[edit]- ^ There are advantages and disadvantages for all medical screening tests. Clinical practice guidelines, such as those for colorectal cancer screening, describe these risks and benefits.[24][25]
References
[edit]- ^ Yerushalmy J (1947). "Statistical problems in assessing methods of medical diagnosis with special reference to x-ray techniques". Public Health Reports. 62 (2): 1432–39. doi:10.2307/4586294. JSTOR 4586294. PMID 20340527. S2CID 19967899.
- ^ Saah AJ, Hoover DR (1998). "[Sensitivity and specificity revisited: significance of the terms in analytic and diagnostic language]". Ann Dermatol Venereol. 125 (4): 291–4. PMID 9747274.
- ^ Parikh R, Mathai A, Parikh S, Chandra Sekhar G, Thomas R (2008). "Understanding and using sensitivity, specificity and predictive values". Indian Journal of Ophthalmology. 56 (1): 45–50. doi:10.4103/0301-4738.37595. PMC 2636062. PMID 18158403.
- ^ a b Altman DG, Bland JM (June 1994). "Diagnostic tests. 1: Sensitivity and specificity". BMJ. 308 (6943): 1552. doi:10.1136/bmj.308.6943.1552. PMC 2540489. PMID 8019315.
- ^ "SpPin and SnNout". Centre for Evidence Based Medicine (CEBM). Retrieved 18 January 2023.
- ^ Mangrulkar R. "Diagnostic Reasoning I and II". Archived from the original on 1 August 2011. Retrieved 24 January 2012.
- ^ "Evidence-Based Diagnosis". Michigan State University. Archived from the original on 2013-07-06. Retrieved 2013-08-23.
- ^ "Sensitivity and Specificity". Emory University Medical School Evidence Based Medicine course.
- ^ Baron JA (Apr–Jun 1994). "Too bad it isn't true". Medical Decision Making. 14 (2): 107. doi:10.1177/0272989X9401400202. PMID 8028462. S2CID 44505648.
- ^ Boyko EJ (Apr–Jun 1994). "Ruling out or ruling in disease with the most sensitive or specific diagnostic test: short cut or wrong turn?". Medical Decision Making. 14 (2): 175–9. doi:10.1177/0272989X9401400210. PMID 8028470. S2CID 31400167.
- ^ Pewsner D, Battaglia M, Minder C, Marx A, Bucher HC, Egger M (July 2004). "Ruling a diagnosis in or out with "SpPIn" and "SnNOut": a note of caution". BMJ. 329 (7459): 209–13. doi:10.1136/bmj.329.7459.209. PMC 487735. PMID 15271832.
- ^ Fawcett T (2006). "An Introduction to ROC Analysis". Pattern Recognition Letters. 27 (8): 861–874. Bibcode:2006PaReL..27..861F. CiteSeerX 10.1.1.646.2144. doi:10.1016/j.patrec.2005.10.010. S2CID 2027090.
- ^ a b Powers DM (2011). "Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness & Correlation". Journal of Machine Learning Technologies. 2 (1): 37–63.
- ^ Gale SD, Perkel DJ (January 2010). "A basal ganglia pathway drives selective auditory responses in songbird dopaminergic neurons via disinhibition". The Journal of Neuroscience. 30 (3): 1027–37. doi:10.1523/JNEUROSCI.3585-09.2010. PMC 2824341. PMID 20089911.
- ^ Macmillan NA, Creelman CD (15 September 2004). Detection Theory: A User's Guide. Psychology Press. p. 7. ISBN 978-1-4106-1114-7.
- ^ Fawcett T (2006). "An Introduction to ROC Analysis" (PDF). Pattern Recognition Letters. 27 (8): 861–874. doi:10.1016/j.patrec.2005.10.010. S2CID 2027090.
- ^ Provost F, Tom Fawcett (2013-08-01). "Data Science for Business: What You Need to Know about Data Mining and Data-Analytic Thinking". O'Reilly Media, Inc.
- ^ Powers DM (2011). "Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness & Correlation". Journal of Machine Learning Technologies. 2 (1): 37–63.
- ^ Ting KM (2011). Sammut C, Webb GI (eds.). Encyclopedia of machine learning. Springer. doi:10.1007/978-0-387-30164-8. ISBN 978-0-387-30164-8.
- ^ Brooks H, Brown B, Ebert B, Ferro C, Jolliffe I, Koh TY, Roebber P, Stephenson D (2015-01-26). "WWRP/WGNE Joint Working Group on Forecast Verification Research". Collaboration for Australian Weather and Climate Research. World Meteorological Organisation. Retrieved 2019-07-17.
- ^ Chicco D, Jurman G (January 2020). "The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation". BMC Genomics. 21 (1): 6-1–6-13. doi:10.1186/s12864-019-6413-7. PMC 6941312. PMID 31898477.
- ^ Chicco D, Toetsch N, Jurman G (February 2021). "The Matthews correlation coefficient (MCC) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation". BioData Mining. 14 (13): 13. doi:10.1186/s13040-021-00244-z. PMC 7863449. PMID 33541410.
- ^ Tharwat A. (August 2018). "Classification assessment methods". Applied Computing and Informatics. 17: 168–192. doi:10.1016/j.aci.2018.08.003.
- ^ Lin JS, Piper MA, Perdue LA, Rutter CM, Webber EM, O'Connor E, Smith N, Whitlock EP (21 June 2016). "Screening for Colorectal Cancer". JAMA. 315 (23): 2576–2594. doi:10.1001/jama.2016.3332. ISSN 0098-7484. PMID 27305422.
- ^ Bénard F, Barkun AN, Martel M, Renteln Dv (7 January 2018). "Systematic review of colorectal cancer screening guidelines for average-risk adults: Summarizing the current global recommendations". World Journal of Gastroenterology. 24 (1): 124–138. doi:10.3748/wjg.v24.i1.124. PMC 5757117. PMID 29358889.
- ^ "Diagnostic test online calculator calculates sensitivity, specificity, likelihood ratios and predictive values from a 2x2 table – calculator of confidence intervals for predictive parameters". medcalc.org.
- ^ Burge C, Karlin S (1997). "Prediction of complete gene structures in human genomic DNA" (PDF). Journal of Molecular Biology. 268 (1): 78–94. CiteSeerX 10.1.1.115.3107. doi:10.1006/jmbi.1997.0951. PMID 9149143. Archived from the original (PDF) on 2015-06-20.
- ^ "GeneMark-ES". Lomsadze A (2005). "Gene finding in novel genomes by self-training algorithm". Nucleic Acids Research. 33 (20): 6494–6906. doi:10.1093/nar/gki937. PMC 1298918. PMID 16314312.
- ^ Korf I (2004). "Gene finding in novel genomes". BMC Bioinformatics. 5: 59. doi:10.1186/1471-2105-5-59. PMC 421630. PMID 15144565.
- ^ Yandell M, Ence D (April 2012). "A beginner's guide to eukaryotic genome annotation". Nature Reviews. Genetics. 13 (5): 329–42. doi:10.1038/nrg3174. PMID 22510764. S2CID 3352427.
Further reading
[edit]- Altman DG, Bland JM (June 1994). "Diagnostic tests. 1: Sensitivity and specificity". BMJ. 308 (6943): 1552. doi:10.1136/bmj.308.6943.1552. PMC 2540489. PMID 8019315.
- Loong TW (September 2003). "Understanding sensitivity and specificity with the right side of the brain". BMJ. 327 (7417): 716–9. doi:10.1136/bmj.327.7417.716. PMC 200804. PMID 14512479.

![{\displaystyle {\begin{aligned}{\text{sensitivity}}&={\frac {\text{number of true positives}}{{\text{number of true positives}}+{\text{number of false negatives}}}}\\[8pt]&={\frac {\text{number of true positives}}{\text{total number of sick individuals in population}}}\\[8pt]&={\text{probability of a positive test given that the patient has the disease}}\end{aligned}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/12ec58e26222c7c528150ce69c86e2aa91ddc4c2)
![{\displaystyle {\begin{aligned}{\text{specificity}}&={\frac {\text{number of true negatives}}{{\text{number of true negatives}}+{\text{number of false positives}}}}\\[8pt]&={\frac {\text{number of true negatives}}{\text{total number of well individuals in population}}}\\[8pt]&={\text{probability of a negative test given that the patient is well}}\end{aligned}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/d48cee2cea0745bcc29d228f8c2783e4cb34547c)











