Molecular biology
| Part of a series on |
| Biology |
|---|
Molecular biology /məˈlɛkjʊlər/ is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.[1][2][3]
Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observations of so-called classical biology, which instead studies biological processes at larger scales and higher levels of organization.[4] In 1953, Francis Crick, James Watson, Rosalind Franklin, and their colleagues at the Medical Research Council Unit, Cavendish Laboratory, were the first to describe the double helix model for the chemical structure of deoxyribonucleic acid (DNA), which is often considered a landmark event for the nascent field because it provided a physico-chemical basis by which to understand the previously nebulous idea of nucleic acids as the primary substance of biological inheritance. They proposed this structure based on previous research done by Franklin, which was conveyed to them by Maurice Wilkins and Max Perutz.[5] Their work led to the discovery of DNA in other microorganisms, plants, and animals.[6]
The field of molecular biology includes techniques which enable scientists to learn about molecular processes.[7] These techniques are used to efficiently target new drugs, diagnose disease, and better understand cell physiology.[8] Some clinical research and medical therapies arising from molecular biology are covered under gene therapy, whereas the use of molecular biology or molecular cell biology in medicine is now referred to as molecular medicine.[citation needed]
History of molecular biology
[edit]

Molecular biology sits at the intersection of biochemistry and genetics; as these scientific disciplines emerged and evolved in the 20th century, it became clear that they both sought to determine the molecular mechanisms which underlie vital cellular functions.[9][10] Advances in molecular biology have been closely related to the development of new technologies and their optimization.[11] Molecular biology has been elucidated by the work of many scientists, and thus the history of the field depends on an understanding of these scientists and their experiments.[citation needed]
The field of genetics arose from attempts to understand the set of rules underlying reproduction and heredity, and the nature of the hypothetical units of heredity known as genes. Gregor Mendel pioneered this work in 1866, when he first described the laws of inheritance he observed in his studies of mating crosses in pea plants.[12] One such law of genetic inheritance is the law of segregation, which states that diploid individuals with two alleles for a particular gene will pass one of these alleles to their offspring.[13] Because of his critical work, the study of genetic inheritance is commonly referred to as Mendelian genetics.[14]
A major milestone in molecular biology was the discovery of the structure of DNA. This work began in 1869 by Friedrich Miescher, a Swiss biochemist who first proposed a structure called nuclein, which we now know to be (deoxyribonucleic acid), or DNA.[15] He discovered this unique substance by studying the components of pus-filled bandages, and noting the unique properties of the "phosphorus-containing substances".[16] Another notable contributor to the DNA model was Phoebus Levene, who proposed the "polynucleotide model" of DNA in 1919 as a result of his biochemical experiments on yeast.[17] In 1950, Erwin Chargaff expanded on the work of Levene and elucidated a few critical properties of nucleic acids: first, the sequence of nucleic acids varies across species.[18] Second, the total concentration of purines (adenine and guanine) is always equal to the total concentration of pyrimidines (cysteine and thymine).[15] This is now known as Chargaff's rule. In 1953, James Watson and Francis Crick published the double helical structure of DNA,[19] based on the X-ray crystallography work done by Rosalind Franklin which was conveyed to them by Maurice Wilkins and Max Perutz.[5] Watson and Crick described the structure of DNA and conjectured about the implications of this unique structure for possible mechanisms of DNA replication.[19] Watson and Crick were awarded the Nobel Prize in Physiology or Medicine in 1962, along with Wilkins, for proposing a model of the structure of DNA.[6]
In 1961, it was demonstrated that when a gene encodes a protein, three sequential bases of a gene's DNA specify each successive amino acid of the protein.[20] Thus the genetic code is a triplet code, where each triplet (called a codon) specifies a particular amino acid. Furthermore, it was shown that the codons do not overlap with each other in the DNA sequence encoding a protein, and that each sequence is read from a fixed starting point. During 1962–1964, through the use of conditional lethal mutants of a bacterial virus,[21] fundamental advances were made in our understanding of the functions and interactions of the proteins employed in the machinery of DNA replication, DNA repair, DNA recombination, and in the assembly of molecular structures.[22]
Griffith's experiment
[edit]
In 1928, Frederick Griffith, encountered a virulence property in pneumococcus bacteria, which was killing lab rats. According to Mendel, prevalent at that time, gene transfer could occur only from parent to daughter cells. Griffith advanced another theory, stating that gene transfer occurring in member of same generation is known as horizontal gene transfer (HGT). This phenomenon is now referred to as genetic transformation.[23]
Griffith's experiment addressed the pneumococcus bacteria, which had two different strains, one virulent and smooth and one avirulent and rough. The smooth strain had glistering appearance owing to the presence of a type of specific polysaccharide – a polymer of glucose and glucuronic acid capsule. Due to this polysaccharide layer of bacteria, a host's immune system cannot recognize the bacteria and it kills the host. The other, avirulent, rough strain lacks this polysaccharide capsule and has a dull, rough appearance.[citation needed]
Presence or absence of capsule in the strain, is known to be genetically determined. Smooth and rough strains occur in several different type such as S-I, S-II, S-III, etc. and R-I, R-II, R-III, etc. respectively. All this subtypes of S and R bacteria differ with each other in antigen type they produce.[6]
Avery–MacLeod–McCarty experiment
[edit]The Avery–MacLeod–McCarty experiment was a landmark study conducted in 1944 that demonstrated that DNA, not protein as previously thought, carries genetic information in bacteria. Oswald Avery, Colin Munro MacLeod, and Maclyn McCarty used an extract from a strain of pneumococcus that could cause pneumonia in mice. They showed that genetic transformation in the bacteria could be accomplished by injecting them with purified DNA from the extract. They discovered that when they digested the DNA in the extract with DNase, transformation of harmless bacteria into virulent ones was lost. This provided strong evidence that DNA was the genetic material, challenging the prevailing belief that proteins were responsible. It laid the basis for the subsequent discovery of its structure by Watson and Crick.
Hershey–Chase experiment
[edit]
Confirmation that DNA is the genetic material which is cause of infection came from the Hershey–Chase experiment. They used E.coli and bacteriophage for the experiment. This experiment is also known as blender experiment, as kitchen blender was used as a major piece of apparatus. Alfred Hershey and Martha Chase demonstrated that the DNA injected by a phage particle into a bacterium contains all information required to synthesize progeny phage particles. They used radioactivity to tag the bacteriophage's protein coat with radioactive sulphur and DNA with radioactive phosphorus, into two different test tubes respectively. After mixing bacteriophage and E.coli into the test tube, the incubation period starts in which phage transforms the genetic material in the E.coli cells. Then the mixture is blended or agitated, which separates the phage from E.coli cells. The whole mixture is centrifuged and the pellet which contains E.coli cells was checked and the supernatant was discarded. The E.coli cells showed radioactive phosphorus, which indicated that the transformed material was DNA not the protein coat.
The transformed DNA gets attached to the DNA of E.coli and radioactivity is only seen onto the bacteriophage's DNA. This mutated DNA can be passed to the next generation and the theory of Transduction came into existence. Transduction is a process in which the bacterial DNA carry the fragment of bacteriophages and pass it on the next generation. This is also a type of horizontal gene transfer.[6]
Meselson–Stahl experiment
[edit]
The Meselson-Stahl experiment was a landmark experiment in molecular biology that provided evidence for the semiconservative replication of DNA. Conducted in 1958 by Matthew Meselson and Franklin Stahl, the experiment involved growing E. coli bacteria in a medium containing heavy isotope of nitrogen (15N) for several generations. This caused all the newly synthesized bacterial DNA to be incorporated with the heavy isotope.
After allowing the bacteria to replicate in a medium containing normal nitrogen (14N), samples were taken at various time points. These samples were then subjected to centrifugation in a density gradient, which separated the DNA molecules based on their density.
The results showed that after one generation of replication in the 14N medium, the DNA formed a band of intermediate density between that of pure 15N DNA and pure 14N DNA. This supported the semiconservative DNA replication proposed by Watson and Crick, where each strand of the parental DNA molecule serves as a template for the synthesis of a new complementary strand, resulting in two daughter DNA molecules, each consisting of one parental and one newly synthesized strand.
The Meselson-Stahl experiment provided compelling evidence for the semiconservative replication of DNA, which is fundamental to the understanding of genetics and molecular biology.
Modern molecular biology
[edit]In the early 2020s, molecular biology entered a golden age defined by both vertical and horizontal technical development. Vertically, novel technologies are allowing for real-time monitoring of biological processes at the atomic level.[24] Molecular biologists today have access to increasingly affordable sequencing data at increasingly higher depths, facilitating the development of novel genetic manipulation methods in new non-model organisms. Likewise, synthetic molecular biologists will drive the industrial production of small and macro molecules through the introduction of exogenous metabolic pathways in various prokaryotic and eukaryotic cell lines.[25]
Horizontally, sequencing data is becoming more affordable and used in many different scientific fields. This will drive the development of industries in developing nations and increase accessibility to individual researchers. Likewise, CRISPR-Cas9 gene editing experiments can now be conceived and implemented by individuals for under $10,000 in novel organisms, which will drive the development of industrial and medical applications.[26]
Relationship to other biological sciences
[edit]
The following list describes a viewpoint on the interdisciplinary relationships between molecular biology and other related fields.[27]
- Molecular biology is the study of the molecular underpinnings of the biological phenomena, focusing on molecular synthesis, modification, mechanisms and interactions.
- Biochemistry is the study of the chemical substances and vital processes occurring in living organisms. Biochemists focus heavily on the role, function, and structure of biomolecules such as proteins, lipids, carbohydrates and nucleic acids.[28]
- Genetics is the study of how genetic differences affect organisms. Genetics attempts to predict how mutations, individual genes and genetic interactions can affect the expression of a phenotype[29]
While researchers practice techniques specific to molecular biology, it is common to combine these with methods from genetics and biochemistry. Much of molecular biology is quantitative, and recently a significant amount of work has been done using computer science techniques such as bioinformatics and computational biology. Molecular genetics, the study of gene structure and function, has been among the most prominent sub-fields of molecular biology since the early 2000s. Other branches of biology are informed by molecular biology, by either directly studying the interactions of molecules in their own right such as in cell biology and developmental biology, or indirectly, where molecular techniques are used to infer historical attributes of populations or species, as in fields in evolutionary biology such as population genetics and phylogenetics. There is also a long tradition of studying biomolecules "from the ground up", or molecularly, in biophysics.[30]
Techniques of molecular biology
[edit]
Molecular cloning
[edit]
Molecular cloning is used to isolate and then transfer a DNA sequence of interest into a plasmid vector.[31] This recombinant DNA technology was first developed in the 1960s.[32] In this technique, a DNA sequence coding for a protein of interest is cloned using polymerase chain reaction (PCR), and/or restriction enzymes, into a plasmid (expression vector). The plasmid vector usually has at least 3 distinctive features: an origin of replication, a multiple cloning site (MCS), and a selective marker (usually antibiotic resistance). Additionally, upstream of the MCS are the promoter regions and the transcription start site, which regulate the expression of cloned gene.
This plasmid can be inserted into either bacterial or animal cells. Introducing DNA into bacterial cells can be done by transformation via uptake of naked DNA, conjugation via cell-cell contact or by transduction via viral vector. Introducing DNA into eukaryotic cells, such as animal cells, by physical or chemical means is called transfection. Several different transfection techniques are available, such as calcium phosphate transfection, electroporation, microinjection and liposome transfection. The plasmid may be integrated into the genome, resulting in a stable transfection, or may remain independent of the genome and expressed temporarily, called a transient transfection.[33][34]
DNA coding for a protein of interest is now inside a cell, and the protein can now be expressed. A variety of systems, such as inducible promoters and specific cell-signaling factors, are available to help express the protein of interest at high levels. Large quantities of a protein can then be extracted from the bacterial or eukaryotic cell. The protein can be tested for enzymatic activity under a variety of situations, the protein may be crystallized so its tertiary structure can be studied, or, in the pharmaceutical industry, the activity of new drugs against the protein can be studied.[35]
Polymerase chain reaction
[edit]Polymerase chain reaction (PCR) is an extremely versatile technique for copying DNA. In brief, PCR allows a specific DNA sequence to be copied or modified in predetermined ways. The reaction is extremely powerful and under perfect conditions could amplify one DNA molecule to become 1.07 billion molecules in less than two hours. PCR has many applications, including the study of gene expression, the detection of pathogenic microorganisms, the detection of genetic mutations, and the introduction of mutations to DNA.[36] The PCR technique can be used to introduce restriction enzyme sites to ends of DNA molecules, or to mutate particular bases of DNA, the latter is a method referred to as site-directed mutagenesis. PCR can also be used to determine whether a particular DNA fragment is found in a cDNA library. PCR has many variations, like reverse transcription PCR (RT-PCR) for amplification of RNA, and, more recently, quantitative PCR which allow for quantitative measurement of DNA or RNA molecules.[37][38]

Gel electrophoresis
[edit]
Gel electrophoresis is a technique which separates molecules by their size using an agarose or polyacrylamide gel.[39] This technique is one of the principal tools of molecular biology. The basic principle is that DNA fragments can be separated by applying an electric current across the gel - because the DNA backbone contains negatively charged phosphate groups, the DNA will migrate through the agarose gel towards the positive end of the current.[39] Proteins can also be separated on the basis of size using an SDS-PAGE gel, or on the basis of size and their electric charge by using what is known as a 2D gel electrophoresis.[40]

The Bradford protein assay
[edit]The Bradford assay is a molecular biology technique which enables the fast, accurate quantitation of protein molecules utilizing the unique properties of a dye called Coomassie Brilliant Blue G-250.[41] Coomassie Blue undergoes a visible color shift from reddish-brown to bright blue upon binding to protein.[41] In its unstable, cationic state, Coomassie Blue has a background wavelength of 465 nm and gives off a reddish-brown color.[42] When Coomassie Blue binds to protein in an acidic solution, the background wavelength shifts to 595 nm and the dye gives off a bright blue color.[42] Proteins in the assay bind Coomassie blue in about 2 minutes, and the protein-dye complex is stable for about an hour, although it is recommended that absorbance readings are taken within 5 to 20 minutes of reaction initiation.[41] The concentration of protein in the Bradford assay can then be measured using a visible light spectrophotometer, and therefore does not require extensive equipment.[42]
This method was developed in 1975 by Marion M. Bradford, and has enabled significantly faster, more accurate protein quantitation compared to previous methods: the Lowry procedure and the biuret assay.[41] Unlike the previous methods, the Bradford assay is not susceptible to interference by several non-protein molecules, including ethanol, sodium chloride, and magnesium chloride.[41] However, it is susceptible to influence by strong alkaline buffering agents, such as sodium dodecyl sulfate (SDS).[41]
Macromolecule blotting and probing
[edit]The terms northern, western and eastern blotting are derived from what initially was a molecular biology joke that played on the term Southern blotting, after the technique described by Edwin Southern for the hybridisation of blotted DNA. Patricia Thomas, developer of the RNA blot which then became known as the northern blot, actually did not use the term.[43]
Southern blotting
[edit]Named after its inventor, biologist Edwin Southern, the Southern blot is a method for probing for the presence of a specific DNA sequence within a DNA sample. DNA samples before or after restriction enzyme (restriction endonuclease) digestion are separated by gel electrophoresis and then transferred to a membrane by blotting via capillary action. The membrane is then exposed to a labeled DNA probe that has a complement base sequence to the sequence on the DNA of interest.[44] Southern blotting is less commonly used in laboratory science due to the capacity of other techniques, such as PCR, to detect specific DNA sequences from DNA samples. These blots are still used for some applications, however, such as measuring transgene copy number in transgenic mice or in the engineering of gene knockout embryonic stem cell lines.[30]
Northern blotting
[edit]
The northern blot is used to study the presence of specific RNA molecules as relative comparison among a set of different samples of RNA. It is essentially a combination of denaturing RNA gel electrophoresis, and a blot. In this process RNA is separated based on size and is then transferred to a membrane that is then probed with a labeled complement of a sequence of interest. The results may be visualized through a variety of ways depending on the label used; however, most result in the revelation of bands representing the sizes of the RNA detected in sample. The intensity of these bands is related to the amount of the target RNA in the samples analyzed. The procedure is commonly used to study when and how much gene expression is occurring by measuring how much of that RNA is present in different samples, assuming that no post-transcriptional regulation occurs and that the levels of mRNA reflect proportional levels of the corresponding protein being produced. It is one of the most basic tools for determining at what time, and under what conditions, certain genes are expressed in living tissues.[45][46]
Western blotting
[edit]A western blot is a technique by which specific proteins can be detected from a mixture of proteins.[47] Western blots can be used to determine the size of isolated proteins, as well as to quantify their expression.[48] In western blotting, proteins are first separated by size, in a thin gel sandwiched between two glass plates in a technique known as SDS-PAGE. The proteins in the gel are then transferred to a polyvinylidene fluoride (PVDF), nitrocellulose, nylon, or other support membrane. This membrane can then be probed with solutions of antibodies. Antibodies that specifically bind to the protein of interest can then be visualized by a variety of techniques, including colored products, chemiluminescence, or autoradiography. Often, the antibodies are labeled with enzymes. When a chemiluminescent substrate is exposed to the enzyme it allows detection. Using western blotting techniques allows not only detection but also quantitative analysis. Analogous methods to western blotting can be used to directly stain specific proteins in live cells or tissue sections.[47][49]
Eastern blotting
[edit]The eastern blotting technique is used to detect post-translational modification of proteins. Proteins blotted on to the PVDF or nitrocellulose membrane are probed for modifications using specific substrates.[50]
Microarrays
[edit]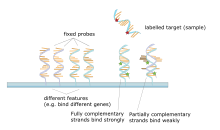
A DNA microarray is a collection of spots attached to a solid support such as a microscope slide where each spot contains one or more single-stranded DNA oligonucleotide fragments. Arrays make it possible to put down large quantities of very small (100 micrometre diameter) spots on a single slide. Each spot has a DNA fragment molecule that is complementary to a single DNA sequence. A variation of this technique allows the gene expression of an organism at a particular stage in development to be qualified (expression profiling). In this technique the RNA in a tissue is isolated and converted to labeled complementary DNA (cDNA). This cDNA is then hybridized to the fragments on the array and visualization of the hybridization can be done. Since multiple arrays can be made with exactly the same position of fragments, they are particularly useful for comparing the gene expression of two different tissues, such as a healthy and cancerous tissue. Also, one can measure what genes are expressed and how that expression changes with time or with other factors. There are many different ways to fabricate microarrays; the most common are silicon chips, microscope slides with spots of ~100 micrometre diameter, custom arrays, and arrays with larger spots on porous membranes (macroarrays). There can be anywhere from 100 spots to more than 10,000 on a given array. Arrays can also be made with molecules other than DNA.[51][52][53][54]
Allele-specific oligonucleotide
[edit]Allele-specific oligonucleotide (ASO) is a technique that allows detection of single base mutations without the need for PCR or gel electrophoresis. Short (20–25 nucleotides in length), labeled probes are exposed to the non-fragmented target DNA, hybridization occurs with high specificity due to the short length of the probes and even a single base change will hinder hybridization. The target DNA is then washed and the unhybridized probes are removed. The target DNA is then analyzed for the presence of the probe via radioactivity or fluorescence. In this experiment, as in most molecular biology techniques, a control must be used to ensure successful experimentation.[55][56]
In molecular biology, procedures and technologies are continually being developed and older technologies abandoned. For example, before the advent of DNA gel electrophoresis (agarose or polyacrylamide), the size of DNA molecules was typically determined by rate sedimentation in sucrose gradients, a slow and labor-intensive technique requiring expensive instrumentation; prior to sucrose gradients, viscometry was used. Aside from their historical interest, it is often worth knowing about older technology, as it is occasionally useful to solve another new problem for which the newer technique is inappropriate.[57]
See also
[edit]References
[edit]- ^ Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P (2014). Molecular Biology of the Cell, Sixth Edition. Garland Science. pp. 1–10. ISBN 978-1-317-56375-4.
- ^ Gannon F (February 2002). "Molecular biology--what's in a name?". EMBO Reports. 3 (2): 101. doi:10.1093/embo-reports/kvf039. PMC 1083977. PMID 11839687.
- ^ "Molecular biology – Latest research and news | Nature". nature.com. Retrieved 2021-11-07.
- ^ Astbury, W. T. (June 1961). "Molecular Biology or Ultrastructural Biology ?". Nature. 190 (4781): 1124. Bibcode:1961Natur.190.1124A. doi:10.1038/1901124a0. ISSN 1476-4687. PMID 13684868. S2CID 4172248.
- ^ a b "Rosalind Franklin: A Crucial Contribution". nature.com.
- ^ a b c d Verma, P. S. (2004). Cell biology, genetics, molecular biology, evolution and ecology. S Chand and Company. ISBN 81-219-2442-1. OCLC 1045495545.[page needed]
- ^ Morange, Michel (2016). "History of Molecular Biology". Encyclopedia of Life Sciences. pp. 1–8. doi:10.1002/9780470015902.a0003079.pub3. ISBN 978-0-470-01617-6.
- ^ Bello, Elizabeth A.; Schwinn, Debra A. (1996-12-01). "Molecular Biology and Medicine: A Primer for the Clinician". Anesthesiology. 85 (6): 1462–1478. doi:10.1097/00000542-199612000-00029. ISSN 0003-3022. PMID 8968195. S2CID 29581630.
- ^ Bynum, William (February 1999). "A History of Molecular Biology". Nature Medicine. 5 (2): 140. doi:10.1038/5498. ISSN 1546-170X.
- ^ Morange, Michel (June 2021). A history of biology. Princeton University Press. ISBN 978-0-691-18878-2. OCLC 1184123419.[page needed]
- ^ Fields, Stanley (2001-08-28). "The interplay of biology and technology". Proceedings of the National Academy of Sciences. 98 (18): 10051–10054. doi:10.1073/pnas.191380098. ISSN 0027-8424. PMC 56913. PMID 11517346.
- ^ Ellis, T.H. Noel; Hofer, Julie M.I.; Timmerman-Vaughan, Gail M.; Coyne, Clarice J.; Hellens, Roger P. (November 2011). "Mendel, 150 years on". Trends in Plant Science. 16 (11): 590–596. Bibcode:2011TPS....16..590E. doi:10.1016/j.tplants.2011.06.006. PMID 21775188.
- ^ "12.3C: Mendel's Law of Segregation". Biology LibreTexts. 2018-07-12. Retrieved 2021-11-18.
- ^ "Mendelian Inheritance". Genome.gov. Retrieved 2021-11-18.
- ^ a b Pray, L (2008). "Discovery of DNA structure and function: Watson and Crick". Nature Education. 1 (1): 100. Retrieved 2024-06-21.
- ^ George., Wolf (2003). Friedrich Miescher: the man who discovered DNA. OCLC 907773747.[page needed]
- ^ Levene, P.A. (1919). "Structure of Yeast Nucleic Acid". Journal of Biological Chemistry. 43 (2): 379–382. doi:10.1016/s0021-9258(18)86289-5. ISSN 0021-9258.
- ^ Chargaff, Erwin (June 1950). "Chemical specificity of nucleic acids and mechanism of their enzymatic degradation". Experientia. 6 (6): 201–209. doi:10.1007/bf02173653. PMID 15421335. S2CID 2522535.
- ^ a b Watson, J. D.; Crick, F. H. C. (April 1953). "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid". Nature. 171 (4356): 737–738. Bibcode:1953Natur.171..737W. doi:10.1038/171737a0. ISSN 1476-4687. PMID 13054692. S2CID 4253007.
- ^ Crick, F. H. C.; Barnett, Leslie; Brenner, S.; Watts-Tobin, R. J. (1961). "General Nature of the Genetic Code for Proteins". Nature. 192 (4809). Springer Science and Business Media LLC: 1227–1232. Bibcode:1961Natur.192.1227C. doi:10.1038/1921227a0. ISSN 0028-0836. PMID 13882203. S2CID 4276146.
- ^ Epstein, R. H.; Bolle, A.; Steinberg, C. M.; Kellenberger, E.; Boy de la Tour, E.; et al. (1963-01-01). "Physiological Studies of Conditional Lethal Mutants of Bacteriophage T4D". Cold Spring Harbor Symposia on Quantitative Biology. 28. Cold Spring Harbor Laboratory: 375–394. doi:10.1101/sqb.1963.028.01.053. ISSN 0091-7451.
- ^ Edgar, Bob (2004-10-01). "The Genome of Bacteriophage T4". Genetics. 168 (2): 575–582. doi:10.1093/genetics/168.2.575. ISSN 1943-2631. PMC 1448817. PMID 15514035.
- ^ Ravenhall, Matt; Škunca, Nives; Lassalle, Florent; Dessimoz, Christophe (May 2015). "Inferring Horizontal Gene Transfer". PLOS Computational Biology. 11 (5): e1004095. Bibcode:2015PLSCB..11E4095R. doi:10.1371/journal.pcbi.1004095. PMC 4462595. PMID 26020646.
- ^ Mojiri, Soheil; Isbaner, Sebastian; Mühle, Steffen; Jang, Hongje; Bae, Albert Johann; Gregor, Ingo; Gholami, Azam; Gholami, Azam; Enderlein, Jörg (2021-06-01). "Rapid multi-plane phase-contrast microscopy reveals torsional dynamics in flagellar motion". Biomedical Optics Express. 12 (6): 3169–3180. doi:10.1364/BOE.419099. ISSN 2156-7085. PMC 8221972. PMID 34221652.
- ^ van Warmerdam, T. "Molecular Biology Laboratory Resource". Yourbiohelper.com.
- ^ van Warmerdam, T. "Molecular biology laboratory resource". Yourbiohelper.com.
- ^ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000). Molecular cell biology (4th ed.). New York: Scientific American Books. ISBN 978-0-7167-3136-8.
- ^ Berg, Jeremy (2002). Biochemistry. Tymoczko, John L.; Stryer, Lubert (5th ed.). New York: W.H. Freeman. ISBN 0-7167-3051-0. OCLC 48055706.
- ^ Reference, Genetics Home. "Help Me Understand Genetics". Genetics Home Reference. Retrieved 31 December 2016.
- ^ a b Tian J, ed. (2013). Molecular Imaging: Fundamentals and Applications. Springer-Verlag Berlin & Heidelberg GmbH & Co. K. p. 542. ISBN 9783642343032. Retrieved 2019-07-08.
- ^ "Foundations of Molecular Cloning - Past, Present and Future | NEB". www.neb.com. Retrieved 2021-11-25.
- ^ "Foundations of Molecular Cloning - Past, Present and Future | NEB". www.neb.com. Retrieved 2021-11-04.
- ^ Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Isolating, Cloning, and Sequencing DNA. Retrieved 31 December 2016.
- ^ Lessard, Juliane C. (1 January 2013). "Molecular cloning". Laboratory Methods in Enzymology: DNA. Vol. 529. pp. 85–98. doi:10.1016/B978-0-12-418687-3.00007-0. ISBN 978-0-12-418687-3. ISSN 1557-7988. PMID 24011038.
- ^ Kokate C, Jalalpure SS, Hurakadle PJ (2016). Textbook of Pharmaceutical Biotechnology. Expression Cloning. Elsevier. p. 125. ISBN 9788131239872. Retrieved 2019-07-08.
- ^ Lenstra, J. A. (July 1995). "The applications of the polymerase chain reaction in the life sciences". Cellular and Molecular Biology (Noisy-Le-Grand, France). 41 (5): 603–614. ISSN 0145-5680. PMID 7580841.
- ^ "Polymerase Chain Reaction (PCR)". National Center for Biotechnology Information. U.S. National Library of Medicine. Retrieved 31 December 2016.
- ^ "Polymerase Chain Reaction (PCR) Fact Sheet". National Human Genome Research Institute (NHGRI). Retrieved 31 December 2016.
- ^ a b Lee, Pei Yun; Costumbrado, John; Hsu, Chih-Yuan; Kim, Yong Hoon (2012-04-20). "Agarose Gel Electrophoresis for the Separation of DNA Fragments". Journal of Visualized Experiments (62): 3923. doi:10.3791/3923. ISSN 1940-087X. PMC 4846332. PMID 22546956.
- ^ Lee PY, Costumbrado J, Hsu CY, Kim YH (April 2012). "Agarose gel electrophoresis for the separation of DNA fragments". Journal of Visualized Experiments (62). doi:10.3791/3923. PMC 4846332. PMID 22546956.
- ^ a b c d e f Bradford, Marion M. (May 1976). "A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding". Analytical Biochemistry. 72 (1–2): 248–254. doi:10.1016/0003-2697(76)90527-3. PMID 942051. S2CID 4359292.
- ^ a b c "Protein determination by the Bradford method". www.ruf.rice.edu. Retrieved 2021-11-08.
- ^ Thomas PS (September 1980). "Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose". Proceedings of the National Academy of Sciences of the United States of America. 77 (9): 5201–5. Bibcode:1980PNAS...77.5201T. doi:10.1073/pnas.77.9.5201. PMC 350025. PMID 6159641.
- ^ Brown, Terry (1993). "Southern Blotting". Current Protocols in Immunology. 6: Unit 10.6A. doi:10.1002/0471142735.im1006as06. PMID 18432697.
- ^ Josefsen, Knud; Nielsen, Henrik (2011). "Northern Blotting Analysis". RNA. Methods in Molecular Biology. Vol. 703. pp. 87–105. doi:10.1007/978-1-59745-248-9_7. ISBN 978-1-58829-913-0. PMID 21125485.
- ^ He SL, Green R (1 January 2013). "Northern blotting". Laboratory Methods in Enzymology: RNA. Vol. 530. pp. 75–87. doi:10.1016/B978-0-12-420037-1.00003-8. ISBN 978-0-12-420037-1. PMC 4287216. PMID 24034315.
- ^ a b Mahmood T, Yang PC (September 2012). "Western blot: technique, theory, and trouble shooting". North American Journal of Medical Sciences. 4 (9): 429–34. doi:10.4103/1947-2714.100998 (inactive 2024-10-31). PMC 3456489. PMID 23050259.
{{cite journal}}: CS1 maint: DOI inactive as of October 2024 (link) - ^ "Western blot | Learn Science at Scitable". www.nature.com. Retrieved 2021-11-25.
- ^ Kurien BT, Scofield RH (April 2006). "Western blotting". Methods. 38 (4): 283–93. doi:10.1016/j.ymeth.2005.11.007. PMID 16483794.
- ^ Thomas S, Thirumalapura N, Crossley EC, Ismail N, Walker DH (June 2009). "Antigenic protein modifications in Ehrlichia". Parasite Immunology. 31 (6): 296–303. doi:10.1111/j.1365-3024.2009.01099.x. PMC 2731653. PMID 19493209.
- ^ "Microarrays". National Center for Biotechnology Information. U.S. National Library of Medicine. Retrieved 31 December 2016.
- ^ Bumgarner, Roger (2013). "Overview of DNA Microarrays: Types, Applications, and Their Future". Current Protocols in Molecular Biology. 101: Unit 22.1. doi:10.1002/0471142727.mb2201s101. PMC 4011503. PMID 23288464.
- ^ Govindarajan R, Duraiyan J, Kaliyappan K, Palanisamy M (August 2012). "Microarray and its applications". Journal of Pharmacy & Bioallied Sciences. 4 (Suppl 2): S310-2. doi:10.4103/0975-7406.100283. PMC 3467903. PMID 23066278.
- ^ Tarca AL, Romero R, Draghici S (August 2006). "Analysis of microarray experiments of gene expression profiling". American Journal of Obstetrics and Gynecology. 195 (2): 373–88. doi:10.1016/j.ajog.2006.07.001. PMC 2435252. PMID 16890548.
- ^ Cheng L, Zhang DY, eds. (2008). Molecular genetic pathology. Totowa, NJ: Humana. p. 96. ISBN 978-1-59745-405-6. Retrieved 31 December 2016.
- ^ Leonard DG (2016). Molecular Pathology in Clinical Practice. Springer. p. 31. ISBN 978-3-319-19674-9. Retrieved 31 December 2016.
- ^ Tian J, ed. (2013). Molecular Imaging: Fundamentals and Applications. Springer-Verlag Berlin & Heidelberg GmbH & Co.K. pp. 550, 552. ISBN 9783642343032. Retrieved 2019-07-08.
Further reading
[edit]- Cohen SN, Chang AC, Boyer HW, Helling RB (November 1973). "Construction of biologically functional bacterial plasmids in vitro". Proceedings of the National Academy of Sciences of the United States of America. 70 (11): 3240–4. Bibcode:1973PNAS...70.3240C. doi:10.1073/pnas.70.11.3240. PMC 427208. PMID 4594039.
- Rodgers M (June 1975). "The Pandora's box congress". Rolling Stone. Vol. 189. pp. 37–77.
- Roberts K, Raff M, Alberts B, Walter P, Lewis J, Johnson A (2002). Molecular Biology of the Cell. Garland Science. ISBN 978-0-8153-3218-3.
External links
[edit] Media related to Molecular biology at Wikimedia Commons
Media related to Molecular biology at Wikimedia Commons

